This section contains quality control plots and statistics for the methylation data.
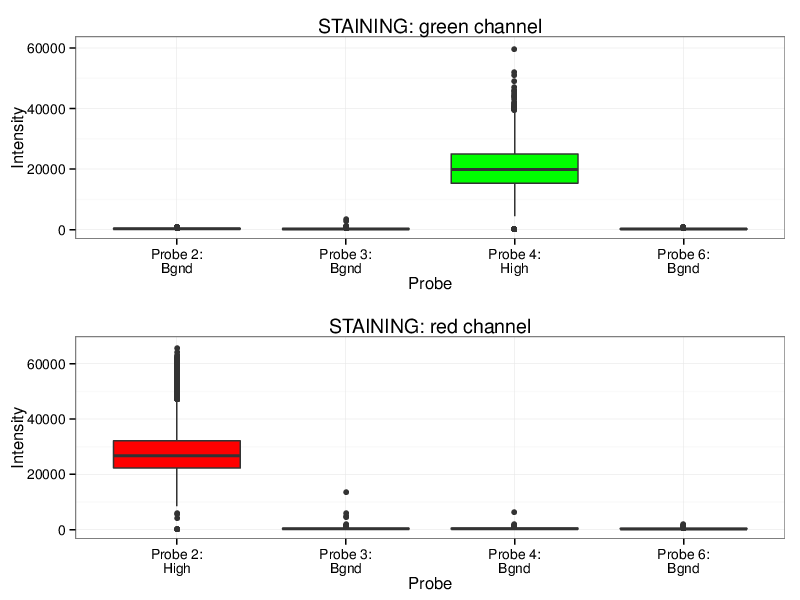
Quality Control Box Plots
Each box plot below shows the signal distribution of quality control probes across all samples. The control box plots are separated by control types. Detailed description of the control probes is given in the RnBeads vignette.
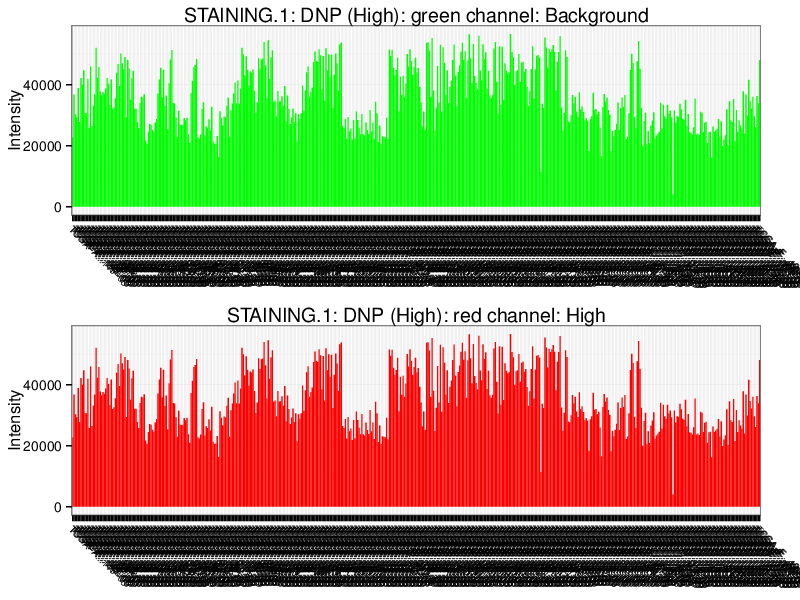
Quality Control Bar Plots
The plots below visualize the exact signal levels at each quality control probe. Note that the scale is not standardized. Background signal is usualy at the level of 1000 to 2000.
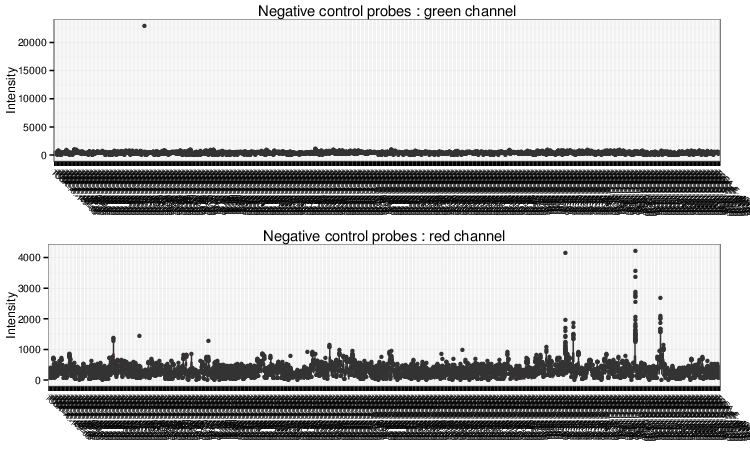
Negative Control Box Plots
Negative control box plots visualize background intensity distributions of all analyzed samples. Samples with skewed distributions and high medians are likely to be of low quality and should be discarded from the analysis.